| 2005 |

|
YEAR BOOK |
Queen's University Belfast
|
The bacterial arms race! – how germs acquire genes for antibiotic resistance
|
The discoveries of antibiotics, vaccines, anaesthetics and asepsis stand as milestones in the history of modern medicine. However with the prevalence of antibiotic resistance amongst microorganisms the future for antibiotic prophylaxis is under threat. Resistance at some level has been recorded against all known antibiotics which raises the questions: Where did resistance originate? and how has it spread so rapidly?
Resistance can even be detected in bacteria from regions where antibiotics have never been used implying that natural resistance predates clinical use. Although antibiotics exert pressure on bacterial populations by selecting for those bacteria with resistance mutations this factor alone does not explain how this resistance has disseminated so rapidly across a wide range of bacterial species. Resistance is believed to involve the conjugation of DNA from one bacterium into another when cells are brought together into temporary contact – a process resulting in horizontal gene transfer.
Research at QUB is investigating the mobility of genetic elements called plasmids – circles of DNA that replicate independently of the chromosome in bacteria. Traditionally it was believed that plasmids could only transfer between closely related bacteria. This hypothesis has now been disproved. Plasmids are capable of transferring between totally unrelated bacteria. Gene transfer between Genera increases the potential for conjugative elements to disseminate antibiotic resistance to many species by tapping into a huge reservoir of potentially resistant genes from bacteria not normally associated with disease.
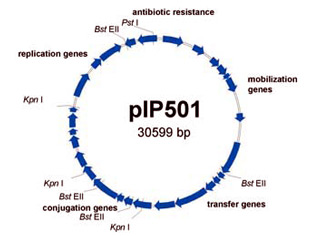
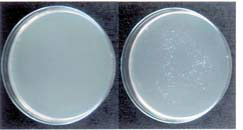
Using streptococcal plasmid pIP501 ( Fig.1 ) as a model, transfer of antibiotic resistance from Enterococcus faecalis to Escherichia coli, two very different bacteria, both of which can inhabit the gastro-intestinal tract, has been detected in the laboratory. It was observed that there was a ten-fold increase in the recovery of chloramphenicol-resistant E. coli only when pIP501 was present during the mating experiments ( Fig.2 ). Although no physical evidence of plasmid transfer could be obtained the antibiotic resistance genes could be detected in the recipient host genome using the polymerase chain reaction.
Contact: Dr Martin Collins, Department of Food Science (Food Microbiology),
Queen's University Belfast;
Tel: 028-90255314; Fax 028-90255009;
E-mail: [email protected]