 Lactobacillus bacteria
The science of biology is being revolutionised by advances in our ability to determine and decipher the DNA molecules that encode the building blocks of all living organisms. The ability to sequence the complete genetic make-up, or genome, of living organisms has only recently been developed and the best-documented application of this technology to date has been the Human Genome Project. However, bacteria represent an attractive target for these technologies due to their small size and less complicated nature. The complete DNA sequences of a number of commercially and medically important bacteria have already been determined. Of particular interest to the dairy industry are members of the Lactic Acid Bacteria (LAB) that impart key flavour, texture and preservation qualities to a variety of fermented foods. The first LAB organism to be completely sequenced was a strain of the cheese starter culture, Lactococcus lactis, but more recent efforts have focused on LAB species that are considered to have health promoting potential such as probiotic Lactobacillus acidophilus and Lactobacillus johnsonii strains.
Lactobacillus bacteria
The science of biology is being revolutionised by advances in our ability to determine and decipher the DNA molecules that encode the building blocks of all living organisms. The ability to sequence the complete genetic make-up, or genome, of living organisms has only recently been developed and the best-documented application of this technology to date has been the Human Genome Project. However, bacteria represent an attractive target for these technologies due to their small size and less complicated nature. The complete DNA sequences of a number of commercially and medically important bacteria have already been determined. Of particular interest to the dairy industry are members of the Lactic Acid Bacteria (LAB) that impart key flavour, texture and preservation qualities to a variety of fermented foods. The first LAB organism to be completely sequenced was a strain of the cheese starter culture, Lactococcus lactis, but more recent efforts have focused on LAB species that are considered to have health promoting potential such as probiotic Lactobacillus acidophilus and Lactobacillus johnsonii strains.
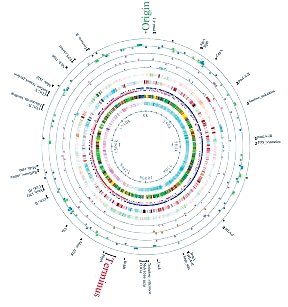 Schematic of a bacterial genome
A major new project is under way at the Dairy Products Research Centre, Moorepark with the objective of determining the complete DNA sequence of a unique cheese making bacterium. The bacterium of interest, Lactobacillus helveticus DPC 4571, was originally isolated from cheese whey and represents one of the most promising candidates for commercial exploitation to emanate from the cheese starter programme at DPRC, Moorepark in recent years. Improvements in the flavour of cheddar cheese manufactured with this bacterium have consistently been observed in both pilot and commercial trials and it has also been used successfully in the development of new cheese varieties. Two particular traits are thought to contribute significantly to the flavour-enhancing abilities of DPC 4571, a potent proteolytic system and the rapid autolysis of the strain in the cheese matrix. Degradation of the major milk protein, casein, by the bacterial proteinase and peptidase enzymes generates important flavour compounds. Moreover, the lysis of the bacterial cell releases these peptidase enzymes into the cheese matrix. Analysis of the DNA sequence of DPC 4571 will facilitate the identification of all the enzymes encoded by the strain and provide a complete picture of the metabolism of this organism.
Schematic of a bacterial genome
A major new project is under way at the Dairy Products Research Centre, Moorepark with the objective of determining the complete DNA sequence of a unique cheese making bacterium. The bacterium of interest, Lactobacillus helveticus DPC 4571, was originally isolated from cheese whey and represents one of the most promising candidates for commercial exploitation to emanate from the cheese starter programme at DPRC, Moorepark in recent years. Improvements in the flavour of cheddar cheese manufactured with this bacterium have consistently been observed in both pilot and commercial trials and it has also been used successfully in the development of new cheese varieties. Two particular traits are thought to contribute significantly to the flavour-enhancing abilities of DPC 4571, a potent proteolytic system and the rapid autolysis of the strain in the cheese matrix. Degradation of the major milk protein, casein, by the bacterial proteinase and peptidase enzymes generates important flavour compounds. Moreover, the lysis of the bacterial cell releases these peptidase enzymes into the cheese matrix. Analysis of the DNA sequence of DPC 4571 will facilitate the identification of all the enzymes encoded by the strain and provide a complete picture of the metabolism of this organism.
The genome sequence of L. helveticus DPC 4571 was determined in collaboration with a commercial biotechnology company and encodes 2.1 million base pairs of nucleotide sequence. Bioinformatic analysis of the sequence data revealed that the genome encodes more than two thousand genes. This data is being systematically 'mined' for genes of potential economic significance using state of the art bioinformatic analysis. The genome of DPC 4571 was initially screened specifically for genes involved in six key metabolic functions that contribute to cheese flavour development, namely proteolysis, amino acid metabolism, lipolysis, exopolysaccharide production, cell lysis and citrate metabolism. From the six functional categories, three have been prioritised and nineteen genes selected for initial investigation based on potential for commercial applications in cheese flavour development. The nineteen genes comprise of bacterial cell lysis genes, lipolytic esterase genes, aminotransferase, proteinase and peptidase genes. The cell lysis genes are important for releasing the bacterial enzymes into the cheese matrix. The aminotransferases, proteinase and peptidases are all involved in the breakdown of the casein and metabolism of its constituent amino acids and, as noted above, these processes contribute significantly to the production of flavour compounds. Degradation of the fat component of milk by lipolytic esterases/lipases is also being increasingly recognised as an important component of cheese flavour development and the selected esterase genes are likely to effect lipolysis of milk fats. The products of all these genes will be investigated with a view to specific commercial applications such as enzyme modified cheese production, generation of bioactive peptides and tools for future starter culture design.
 Swiss cheese
Additional beneficial outputs from the complete genome sequence will be the availability of the data for the development of functional genomic technologies, such as DNA microarrays, and for comparative genomics with other commercially important LAB. All of the currently available Lactobacillus genomes are from strains isolated from the gasterointestinal tract and valued for their probiotic qualities rather than the manufacture of cultured dairy food products. Comparison of the sequenced probiotic lactobacilli and the DPC 4571 cheese culture should provide insight into the flavour-enhancing abilities of DPC 4571 and the mechanisms of intestinal colonisation and health promotion of the probiotic strains. This landmark project is key to building a critical mass in genomics of starter cultures and will place Moorepark at the forefront of international developments in the rapidly evolving area of applying genomics to traditional and innovative food systems.
Swiss cheese
Additional beneficial outputs from the complete genome sequence will be the availability of the data for the development of functional genomic technologies, such as DNA microarrays, and for comparative genomics with other commercially important LAB. All of the currently available Lactobacillus genomes are from strains isolated from the gasterointestinal tract and valued for their probiotic qualities rather than the manufacture of cultured dairy food products. Comparison of the sequenced probiotic lactobacilli and the DPC 4571 cheese culture should provide insight into the flavour-enhancing abilities of DPC 4571 and the mechanisms of intestinal colonisation and health promotion of the probiotic strains. This landmark project is key to building a critical mass in genomics of starter cultures and will place Moorepark at the forefront of international developments in the rapidly evolving area of applying genomics to traditional and innovative food systems.
Contact: Dr. Michael Callanan,
Dr. Tom Beresford,
Dr. Paul Ross;
Tel: 025 42610;
E-mail:
[email protected]
|



