| 2004 |

|
YEAR BOOK |
University College Dublin
|
Computers in Biology – no more experiments?
|
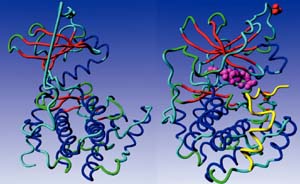
At present, time-consuming and expensive experimental studies comprise the overwhelming majority of studies on enzymes, and large amounts of time and money can therefore be saved if computational methods were capable of calculating the characteristics of enzymes. The characteristics of enzymes are best predicted when the 3D-structure of the enzyme is available, and consequently we are developing algorithms for analysing enzyme 3D-structures.
The development of accurate computational algorithms for the study of enzymes must take place in conjunction with experimental work to validate the theoretical predictions. We are concentrating our efforts in three areas: the protein kinase activation process, the pH-dependence of enzyme catalysis, and the importance of thermal motions for enzyme activity. All three projects aim to improve our ability to substitute experiments with theoretical calculations and, to achieve this goal, we carefully evaluate our theoretical predictions by performing wet-lab experiments.
The project concerning the activation process of the protein kinases (PKs) is of particular importance, since the protein kinases are enzymes that are involved in numerous diseases and, most prominently, in a number of cancers. A computational model that describes the PK activation process accurately will allow for significant advances in the development of drugs for treating cell signalling disorders and cancers.
Contact: Dr Jens Erik Nielsen, Department of Biochemistry, University College Dublin, Dublin 4;
Tel: +353 1 716 6724; E-mail: [email protected] ; Web: http://enzyme.ucd.ie