| 2004 |

|
YEAR BOOK |
Dublin City University
|
Exploiting bacterial genomics for the development of novel antibiotics
|
Critical challenges for modern medicine are posed by the continuing emergence of antibiotic resistance and the preponderance of antibiotic resistant bacteria, particularly in hospital environments. Novel antibiotics, coupled to the management of antibiotic use and diligence in the operation of routine hospital procedures, must all play a role in the control of resistant organisms. The development of novel antibiotics is dependent to a large extent on the identification of new targets against which antibiotics can act in bacterial cells. The completion of almost two hundred bacterial genome sequences, including those of many pathogens, has made available a vast resource from which it will be possible to identify novel targets for antibiotics. One approach to using this resource is to detect the myriad of interactions between encoded proteins and to design or select inhibitors of key interactions. The inhibitors may become lead molecules for the design and synthesis of novel antibiotics.
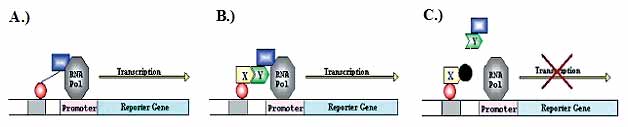
The detection of protein interactions can be undertaken using a variety of physical and genetic techniques. A powerful genetic technique is the two hybrid system, which is demonstrated in panels A and B. The technique involves amplifying segments of the genome and constructing fused genes by DNA cloning. The technique is based on a protein that binds DNA and switches on (activates) the expression of a reporter gene (the product of which can be easily recognised) through the action of two domains within the protein. One domain (domain 1. See below) binds the protein to the DNA and the second (2) directs the activation of the reporter gene. Even when they are separated, these two domains can function, but can only direct the activation if close together. The technique involves fusing one domain to protein X (3) and the second to protein Y (4). If proteins X and Y interact, then the domains are brought sufficiently close together to switch on the reporter, and the interaction can be recognised. By repeating the test for interactions between many of the proteins encoded in a genome, protein interaction maps can be established.

|

|

|

|

|
Contact: Dr. Michael O'Connell, School of Biotechnology, Dublin City University, Glasnevin, Dublin 9; Tel: (01) 7005318; E-mail: [email protected]