| 2004 |

|
YEAR BOOK |
Institute of Technology Carlow
|
GM-Rhizoremediation – Developing sustainable biological solutions for the treatment of soil pollutants
|
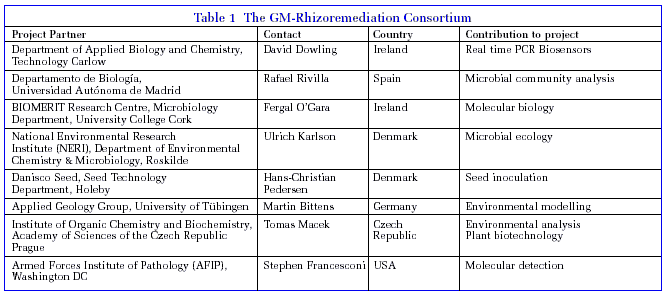
In a second, complementary approach, molecular detection methods for tracking degrader bacteria in the environment will be developed to detect and quantify the GM strains under environmental conditions, and to monitor effects on microbial community structure in the polluted soil. Mathematical environmental modelling of the process and a detailed risk assessment is also under development.
Single cell biosensors
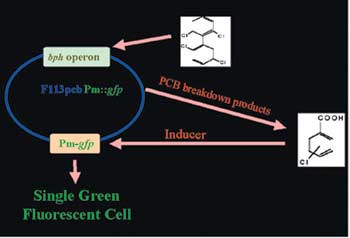
One of the most exciting aspects of the project is the development of integrated biosensor strains to facilitate in situ monitoring of PCB remediation (Figure 1). The system developed allows the detection of bio-available PCBs at levels as low as 100nM. In particular, these tools will allow efficacy monitoring of bioremediation inoculants in real situations. The findings will lead to a better understanding of those measures to be applied for optimisation of phytobioremediation technology.
Molecular detection systems
Novel molecular tools have been developed for the identification and unambiguous quantification of genetically modified (GM) bacterial strains in the environment. In addition, the impact of the inoculant on microbial community composition is being investigated by molecular approaches. Extensive testing of the system in willow plants (Figure 2) and other plant species is near completion. The role of plant in addition to bacterial enzymes is also being assessed. A detailed impact assessment on the use of the GM strains has been undertaken and modelled. We are currently preparing guidelines on the safe use, advantages and limitations of this bioremediation technology.

Future directions
Although working with a model system, the project has generated results that are of general interest that can be applied to bioremediation and environmental protection. These include:
-
Characterised rhizosphere/soil strain(s) to degrade PCBs: principle can be extended to other target compounds
-
Inoculant technologies to introduce strains to the contaminated soil/ rhizosphere environment
-
Biosensor-functionality of strains for monitoring
in situ
activity: could be extended to other compounds e.g. MTB, TBT
-
High-level gene expression system for
Pseudomonas
(based on
Sinorhizobium
nod Box)
Contact: Dr David Dowling, Project Co-ordinator, Institute of Technology Carlow;
E-mail: [email protected] ; Web: http://www.itcarlow.ie/gmrhizo/index.html