| 2006 |

|
YEAR BOOK |
Letterkenny Institute of Technology
|
Pattern Recognition and Machine Learning Research Group at Letterkenny Institute of Technology
|
The electronic voting project is an InterTradeIreland FUSION funded project with OPT2VOTE, the Derry-based software development company specialising in the delivery of e-enabled elections. It is noteworthy that the systems developed by OPT2VOTE for British national elections use normal paper ballot papers, hence an audit trail is assured and manual checking can be carried out at any stage.
Previous projects undertaken by members of the group include inspection of denim fabric, land-use mapping using satellite images, and financial prediction. A number of biomedical imaging projects are in the proposal stage.
The objective of the shellfish recognition work is to help automate the prediction of the optimum time for harvesting shellfish seed; the collection of natural shellfish spat or seed as a source of raw material is a highly valuable shellfish aquaculture industry.
However, due to the time consuming and expertise intensive nature of the task (hours spent on every sample), automated analysis and identification is desirable. In addition, we feel that the techniques being developed will be applicable to a wide range of microscopy work and general machine vision, notably medical applications.
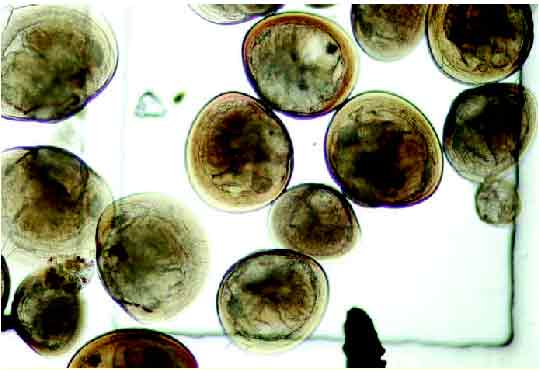

In many cases we are presented with occluding background objects or other larvae, Figure 2(b). Here the edges are faint edges, Figure 2(c), and overlapping; hence boundary edges must be tracked.

Contact: Jonathan Campbell, Department of Computing, Letterkenny Institute of Technology.
E-mail: [email protected]