 Gene expression has become a
powerful tool for determining the spatial and temporal molecular order of biological responses and cellular behaviour. A number of methods exist for detecting and/or isolating genes, which are differentially expressed between two tissues, or the same cell type exposed to more than one environment. Representational difference analysis (RDA) and differential display have gained support for open screening of gene expression, while one expects array analysis to predominate in the future when genome completion and cost factors are fully addressed.
Gene expression has become a
powerful tool for determining the spatial and temporal molecular order of biological responses and cellular behaviour. A number of methods exist for detecting and/or isolating genes, which are differentially expressed between two tissues, or the same cell type exposed to more than one environment. Representational difference analysis (RDA) and differential display have gained support for open screening of gene expression, while one expects array analysis to predominate in the future when genome completion and cost factors are fully addressed.
Subtractive hybridisation offers advantages over the former two in that it reduces the need for large scale sequencing by removing (or subtracting) complimentary DNAs (cDNAs) common to both control and treated samples. The method is used to selectively amplify differentially expressed cDNA fragments and simultaneously suppress non-target DNA amplification, allowing up to a 5000 fold enrichment of rare cDNA corresponding messenger RNAs (mRNAs).
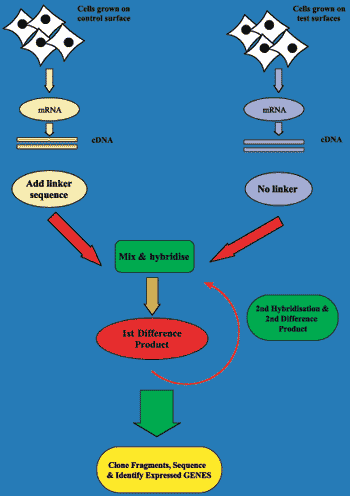 Simplified schematic of Subtractive Hybridisation Methodology
In the Centre for Biopolymer & Biomolecular Research (CBBR) in AIT we have been interested in determining the pattern of gene expression in a liver and vascular cell exposed to a number of different polymeric supports used in biomedical applications. Synchronised human HepG2 cells, human umbilical vein endothelial cells, and human microvascular endothelial cells were grown on a number of collagen coated polymers - aliphatic polycarbonates, aromatic polyethers, a polysiloxane and a control surface. RNA was isolated and cDNA generated. Subtractive hybridisation employs special four cutter restriction enzymes which produce adhesive end ligated adapter molecules and, after two rounds of hybridisation, only differentially expressed fragments spanned by two different adapters are capable of supporting amplification by polymerase chain reaction (PCR). These products are being cloned, sequenced and the gene of origin identified.
Simplified schematic of Subtractive Hybridisation Methodology
In the Centre for Biopolymer & Biomolecular Research (CBBR) in AIT we have been interested in determining the pattern of gene expression in a liver and vascular cell exposed to a number of different polymeric supports used in biomedical applications. Synchronised human HepG2 cells, human umbilical vein endothelial cells, and human microvascular endothelial cells were grown on a number of collagen coated polymers - aliphatic polycarbonates, aromatic polyethers, a polysiloxane and a control surface. RNA was isolated and cDNA generated. Subtractive hybridisation employs special four cutter restriction enzymes which produce adhesive end ligated adapter molecules and, after two rounds of hybridisation, only differentially expressed fragments spanned by two different adapters are capable of supporting amplification by polymerase chain reaction (PCR). These products are being cloned, sequenced and the gene of origin identified.
Preliminary analysis has indicated that cell surface exposure influences the pattern of expression of many genes, including those implicated in cell death, heat shock responses, extracellular matrix synthesis, and this expression pattern is cell type and microenvironment dependent.
We acknowledge support of the Higher Education Authority Programme for Research in Third Level Institutions.
Contact: Dr Paul Tomkins
(Bioserv Ltd/CBBR),
School of Science, Athlone Institute of Technology, Athlone, Co Westmeath, Ireland.
Tel: +353 (0) 902 24453
Fax: +353 (0) 902 24492.
|


