| 2003 |

|
YEAR BOOK |
University of Limerick
|
Structural dissection of bacterial virulence
|
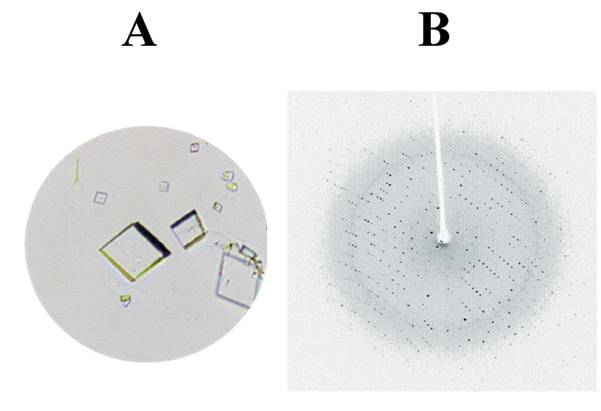
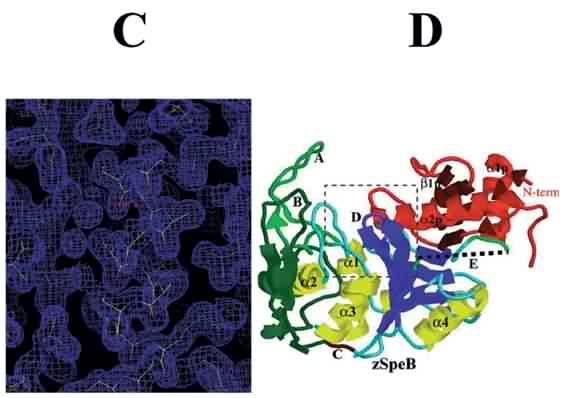
See below for C. and D.
During infection, the bacterial cells produce a variety of agents that are detrimental to the human host. Some of these agents are proteins that aid the bacterial cell to adhere to and/or invade human tissue, or are involved in avoiding detection by the host immune system; others are involved in tissue destruction. The purpose of our studies is to design specific inhibitors targeting bacterial factors and develop novel approaches in combating disease. Using a combination of molecular biological, protein chemistry and structural biology, studies on proteins involved at various stages of the pathogenic process are being undertaken at the University of Limerick.
Solution of protein structure was achieved using X-ray crystallographic methods. The naturally occurring precursor form of the Streptococcal cysteine proteinase, SpeB, was solved at atomic level resolution, which provided a 3D image of the protein molecule. Information provided by analysis of the structure showed that the propeptide-protease interaction was different from those observed in related eukaryotic proteins. Furthermore, the fine detail architecture of the SpeB active site is different. These two findings have allowed development of specific inhibitors of the SpeB protease. It has also allowed structure/ function analysis of key residues involved in stability of the zymogen, a naturally inhibited precursor of an enzyme (e.g. protease), and those contributing to the active site cleft. These studies provide information which drive inhibitor design experiments.
Contact: Dr. Jakki Cooney; Dr. Todd Kagawa; Department of Chemical and Environmental Sciences, University of Limerick. E-mail: [email protected] ; [email protected]