| 2002 |

|
YEAR BOOK |
Health Research Board & University College Cork
|
Identifying genes expressed by Listeria during infection
|
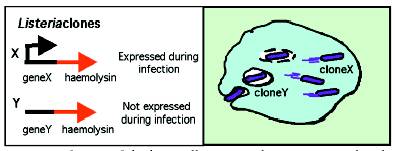
The foodborne pathogen Listeria monocytogenes analysed throughout this study is commonly used as a model for the study of intracellular parasitism. During disease causation, the bacterium invades host cells and is immediately enclosed within the host cell phagosome, a hostile compartment intended to kill invading bacteria. However, Listeria are capable of producing a protein haemolysin that ruptures the host cell phagosome, releasing the bacterium into the host cell where bacterial growth can occur unchecked by the immune system. Listeria bacteria that fail to produce haemolysin cannot grow within host cells, and fail to survive and induce disease in mice.
In addition to expressing the gene encoding haemolysin, it is known that invading Listeria trigger a number of other bacterial genes involved in invasion and survival within the host cell. The current project developed a novel in vivo expression technology (IVET)-based system to detect genes induced by Listeria during infection. The system involved uncoupling haemolysin expression from normal control mechanisms by producing engineered Listeria cells in which the haemolysin gene is under the influence of random genes (see Figure). This bank of random clones was used to infect host cells. Listeria clones of interest were those that survived host cell infection, as these represent fusions of the haemolysin gene to genes expressed during infection.
Using this technology, a number of genes were identified that are preferentially induced by Listeria monocytogenes during host cell infection. These genetic loci include genes with a role in general bacterial metabolism and gene regulation. Furthermore, this system was used to demonstrate that the stress adaptation system GroESL is actively induced by Listeria during infection. It is envisaged that further utilisation of this system will help elucidate the complexities of bacterial gene expression during the infectious process and ultimately inform the design of vaccines and therapeutic strategies against intracellular pathogens.
Cormac Gahan was funded by a Health Research Board Post-Doctoral Research Fellowship.
Contact: Dr Cormac Gahan; E-mail: [email protected]